CRISPR / Cas9
What started as research on bacterial defense against invading foreign DNA resulted in the development of one of the sharpest tools in molecular biology: the CRISPR/Cas9 genetic scissors. This unique molecular tool can be used to make precise cuts in genetic material and enables scientist to easily create changes in the code of life.
Crispr/Cas9 – A manifold tool for genome editing
Since its discovery by Emmanuelle Charpentier and Jennifer A. Doudna, RNA-guided CRISPR/Cas9 system has been adopted as tool to modify genomes of cultured cells, animals as well as plants. Today, CRISPR/Cas9 toolkit is widely used for gene editing, not only in basic science, but also in industry in different fields such as biotechnology and agriculture.
Compared to the classical methods of gene engineering, gene editing is significantly safer as scientists can decide the exact location of the DNA modification via a guiding RNA sequence enabling breakthroughs in the development of therapeutics. Here are some examples for applications of gene editing.
CRISPR/Cas9 in Gene therapy
CRISPR cas is a tool that helps researchers to better understand genetic diseases by exploring the function of genes and with that also their importance in the progress of disease. Building on this, in gene therapies, basically a sick gene is exchanged for a healthy gene allowing a new approach to treat cancer and chronic diseases such as Parkinson.
In September 2020, a gene therapy approach (VY-AADC) showed first improvements in motor functions in patients with Parkinson´s. This approach could potentially facilitate the symptoms of Parkinson´s disease by increasing the levels of dopamine directly in the areas where it is mostly needed.
CRISPR/Cas9 for Agricultural improvement
The climate change poses new challenges for agriculture. Regional plants have to withstand more and more harsh conditions and drought. Under controlled circumstances, scientists are able to improve agriculture by giving plants characteristics to survive in more harsh living conditions. This is possible because with CRISPR, the cleavage of DNA happens precisely at only one predetermined position in the genome compared to classical methods in gene engineering.
CRISPR/Cas9 for SARS-CoV2 detection
Recently, US scientists at UC Berkeley found a new and fast way to directly detect SARS-CoV-2 using an amplification-free CRISPR-Cas13a-based mobile phone assay from nasal swab RNA extracts.
How does the CRISPR/Cas9 toolkit work?
What set the CRISPR/Cas9 toolkit apart from other genome engineering technologies, is its high diversity, modularity, and efficiency. Its most striking advantage is the simplicity by which a gene can be targeted for cleavage in nearly any organism in vitro and in vivo. After cleavage, the targeted gene can be further modified resulting in the respective gene with a deletion, site-directed mutation or a DNA insertion.
Components for DNA cleavage via CRISPR/Cas9
In order to cleave DNA, three elements are essential:
CRISPR-RNA (crRNA)
Cas9 nuclease
trans-activating CRISPR-RNA (tracrRNA)
Explained in detail
1. CRISPR-RNA
The term CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats and was introduced to describe well preserved repetitive DNA sequences in bacteria. These repeats occur in clusters and are separated by unique intervening sequences. The transcription of CRISPR provides long RNA molecules (precursor CRISPR-RNAs, crRNAs), which are subsequently processed by cleavage to yield small CRISPR-RNAs (crRNAs).
In common used CRISPR/Cas9 systems, crRNAs are complementary to the target DNA sequence and show a length of about 20 nucleotides. The target sequence is also named protospacer and must finish with a protospacer adjacent motif (PAM) of the sequence GG or AG at the 3′ end for binding of the nuclease.
CrRNA is required to bind to the target DNA, which is cut by Cas9 when tracrRNA adjusts the nuclease. The final result of Cas9-mediated DNA cleavage is a double-strand break within the target DNA.
2. Cas9 nuclease
In addition to the CRISPR sequences, special genes were discovered always adjacent to CRISPRs. They were called CRISPR-associated genes, encoding cas proteins with helicase and nuclease activity. By now, many different Cas protein subfamilies were described and divided in two classes based on the formation of the complex for antiviral defense. In the Class 1 systems, specialized Cas proteins assemble into a large CRISPR-associated complex providing a cascade activity for DNA cleavage. In contrary, Class 2 systems, such as the Cas9 nuclease, are simpler and contain a single multidomain crRNA-binding protein for similar cleavage activity.
3. tracrRNA
The sequence of the before mentioned CRISP repeats, was unexpectedly found 210 bp upstream of the CRISPR locus on the opposite strand. The transcript was named trans-encoded CRSPR-RNA and contained a stretch of 25 nucleotides almost perfect complementary to the repeat regions of the CRISPR sequence. This RNA has two important functions. First, it can trigger pre-crRNA processing to provide the crRNA for the DNA and Cas interaction. Its second function was discovered by Emmanuelle Charpentier and Jennifer A. Doudna. The two scientists demonstrated that this RNA activated the DNA cleavage by Cas9 as well and was thus the last essential component to be found to form the CRISPR/Cas9 system. Typically, both RNA components, crRNA and tracrRNA, are fused together to form the single-guide RNA molecule (sgRNA).
Today the CRISPR/Cas9 system is delivered as vector encoding the Cas9 nuclease and an engineered sgRNA, enabling scientists with a single transcript to cleave any double-stranded DNA sequence at will.
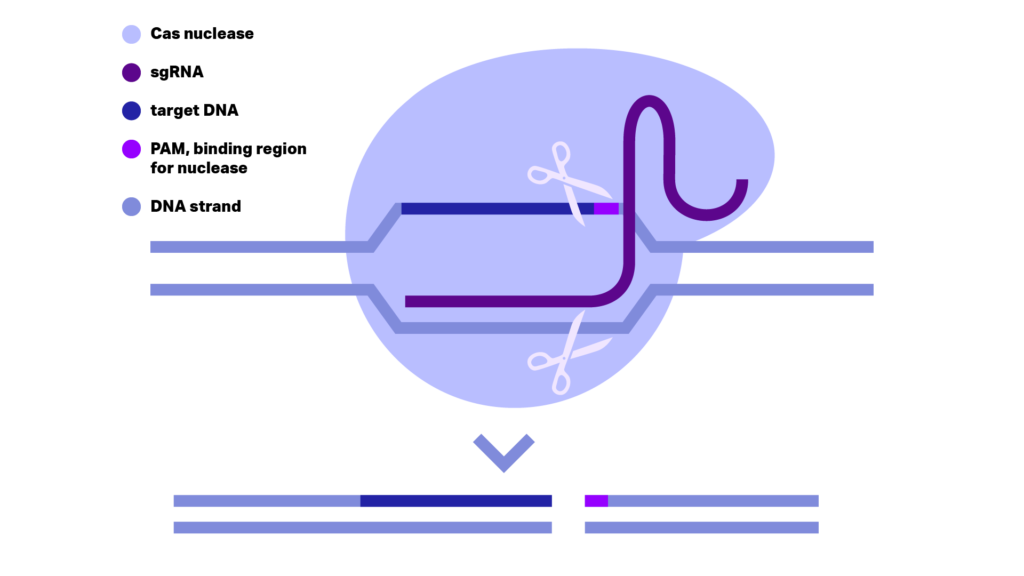
Figure 1: DNA cleavage by CRISPR/Cas9 scissor. sgRNA consisting the crRNA and trcrRNA positioned and activates the Cas9 nuclease on the genome DNA. The correct position of Cas9 is given through crRNA binding on the DNA strand complementary to the target DNA and PAM. Close to PAM the two nuclease domains, RuvC1 and HNH, cut respectively each one of the DNA strands generating a double stranded break
.
Genome Editing using CRISPR/Cas System
The Cas9 induced cleavage allows subsequently precise modifications on the DNA. There are a lot of different possibilities to modify the target DNA. Some examples are described below.
1. Generating knockout phenotypes
The CRISPR/Cas9 induced double-strand break is repaired by the non-homologous end joining (NHEJ) pathway causing small nucleotide insertions or deletions at the target site. These modifications result ideally in a loss-of-function mutation within the targeted gene.
2. Adding a new gene into genome
After CRISPR/Cas9 induced double-strand break, a piece of DNA can be used as a template for homology-directed repair (HDR) resulting in the editing this template into the genome.
3. Introducing site directed point-mutations using base editors
Cas9 can be modified so that the nuclease activity is blocked or only breaks one strand. Such modified Cas9 are additionally fused to other enzymes such as a cytosine deaminase to form a base editor. This Cas mutant can convert cytidine to uridine within a small editing window near the PAM site. DNA repair then converts a guanine–uracil (G–U) intermediate to an adenine–thymine (A–T) base pair.
What are challenges of the CRISPR/Cas9 toolkit?
At the beginning…
the top challenge of using CRISPR/Cas9 system for gene editing was to bring it into different cells and to transcript the nuclease and sgRNA. The components of the CRISPR/Cas system are typically encoded on large extrachromosomal expression vectors with 9–19 kb (kilobase).
Due to the large size of these vectors, the transfection into the cells was inefficient or caused cell death. Improvement in cell transfection methods overcame this challenge allowing the use of single vector transfection into most standard cell lines and thus enabling the extensive application of this technology.
Today…
the major challenge for the application of the CRISPR/Cas9 system is to find the right balance between cleavage efficiency and off-target effects.
Off-target effects arise when genomic cleavage takes place at unintended sequences inducing mutations at sites other than the intended on-target site. These sequences differ by only a few nucleotides from the target DNA sequence and need to be considered when designing a sgRNA for gene editing.
For therapeutic use…
these off-target effects remain a major problem limiting the application of the CRISPR/Cas9 toolkit. Therefore, several approaches have been studied to reduce them without losing cleavage efficiency.
Most of these approaches focus on either the adjusting the sgRNA or the nuclease activity:
Nuclease-based approaches
a. Two separate cuts for improved specificity
In case of the nuclease, CRISPR specificity is improved by using Cas9 nickase mutants or dimeric Cas9 proteins complexed with pairs of sgRNAs. Thus, two nucleases targeting opposite DNA strands are required to generate a double-strand break within the target DNA. The requirement of two close cleavages introduce a further checkpoint for target recognition and thus dramatically increases target specificity. However, the gene delivery for more than one CRISPR/Cas9 system might be challenging for some cell lines.
b. Improvement of cleavage mechanism
Another Cas-based approach is to improve cutting efficiency and specificity through rational mutagenesis providing high fidelity Cas9 variants. This requires intensive studies on the mechanisms by which Cas proteins bind and cleave target DNA.
sgRNA-based approaches
In addition to modifying the nuclease, off-target effect can be reduced by changing the sequence or the length of the gRNA.
a. Strategic sgRNA
One common approach in that regard is the design of strategic sgRNA. Several potential sgRNAs can be utilized to achieve a certain editing. For the selection of the optimal target site and design of corresponding sgRNAs, numerous computational or web-based tools have been developed predicting cleavage efficiency and specificity. The applied models are based on either empirical data or trained by experimental datasets. Future development of these computational tools depends on the availability of extensive datasets for the different experimental conditions and cell lines.
b. Truncated sgRNA
Beside the sequence also changing the length affects the specificity and the off-target effect. Typically, specificity is improved by elongating the sgRNA to involve adjacent genes for the target recognition.
However, researchers recently demonstrated that shorter sgRNAs with only 17 or 18 nucleotides in length allowed specific cleavage, while off-target effect was reduced compared to full-length sgRNAs. In fact, even when the truncated sgRNAs differed from an off-target site by only one or two mismatches, hardly any unintended mutation occurred.
How can Kilobaser help within the CRISPR/Cas 9 technology?
CRISPR/Cas offers a lot of valuable benefits. In order to use these benefits wisely and safely, researchers need to correctly control the target where exactly genomic edits are introduced. This requires optimized gRNA and nuclease activity.
All modifications on a component of the CRISPR/Cas9 system requires customized oligos such as standard DNA primers and DNA coding for new proteins, including mutations.
With Kilobaser all these tools can be prepared directly in the lab in the desired DNA sequence. Especially 20mer crRNAs, which needs to be optimized on a regular basis for the CRISPR technology. crRNA is essential for the position and specificity of DNA cleavage. Using Kilobaser different variations of crRNA can be synthesized fast and whenever they are needed. This would accelerate the optimization of crRNA immensely as different variations can be tested in a noticeably short time. In summary, having a Kilobaser in your own lab would make the whole optimization process truly faster and more efficient.
This allows life science professionals to jump directly in experiments to evaluate the best sequence for their application. With Kilobaser the synthesis of a 25mer custom oligo takes less than 2 hours. For applications like PCR and qPCR Kilobaser DNA primers are ready to use without any additional purification which allows you to do multiple experiments in a day.
Our expertise in chemical oligo synthesis and our innovative spirit makes us the ideal research partner to develop new modifications – chemically modified DNA, such as fluorescence probes for targeting mismatches for detecting off-target cleavages.
Want to read more exciting articles in the future?
Are you interested in our new products and don’t want to miss a thing? Subscribe to our newsletter and stay up to date on what’s going on!

Share this article: