Colony PCR – Application Note
Using the detection of Bacterial Artificial Chromosome (BAC) in E. coli as an example, we performed a colony PCR followed by agarose gel extraction. The DNA primers for this experiment were prepared using Kilobaser personal DNA & RNA synthesizer and compared with DNA primers ordered from Eurofins.

Colony PCR to detect BAC in E. coli
Introduction
Bacterial artificial chromosomes
Plasmids are commonly used to transfect DNA fragments into bacteria. However, the use of plasmids reaches its limits with large DNA fragments starting at a length of about 15,000 bp. In contrast to plasmids, BACs, such as pBelobac11, can hold DNA fragments up to 300,000 bp in size.
This method was used, for example, in the human genome project to sequence the entire DNA of a human being. For this purpose, the human DNA was cut into random parts and incorporated into BACs. Since the BACs contain conserved DNA fragments, the detection of these fragments after transfection into bacteria is easy. DNA was extracted from the bacterial colonies containing BACs and Sanger sequenced.
Colony PCR
Colony PCR bypasses the often critical process of DNA or RNA extraction from bacteria. This provides a faster and less error-prone alternative to detect and amplify a specific DNA or RNA fragment.
In colony PCR, a single bacterial colony is sterilely diluted in water and an aliquot of it is added directly to the PCR reaction. Since the bacterial cell wall denatures during the usual thermal cycles of PCR, this type of PCR has no disadvantage over PCR with previously extracted DNA or RNA in terms of detection and amplification of gene fragments.
KILOBASER DNA synthesis
Kilobaser personal DNA & RNA synthesizer is a benchtop sized micro column synthesizer. Its technology is based on a reagent cartridge system coupled with microfluidic chip.
Standard DNA primers do not need to be purified after synthesis with Kilobaser. Due to the low consumption of reagent volume, the salt residues are negligible. The primers are dissolved in water after synthesis and can be used immediately.
For the synthesis of the DNA primers used in this application, one Kilobaser standard DNA cartridge and 6 Kilobaser standard chips were used.

Material and Methods
Preparation of DNA primer
To verify that the bacteria still contain the bacterial artificial chromosome, specific primers were synthesized with Kilobaser personal DNA synthesizer. A standard reagent cartridge and 6 standard microfluid chips were used.
hV-UL30-FP, ATCAACTTCGACTGGCCCTT
hV-UL30-RP, CCGTACATGTCGATGTTCAC
gC-FP, TCACCATGGAGTTTACGGGC
gC-RP, GTACTCGGTCTGCTCGTACG
UL6-FP, GACCTCAGCAAGTCCATGGA
UL6-RP, GGCGCAGAATCTTGAAGCAG
After synthesis the DNA primers were resuspended in 30 µL nuclease free water by vortexing for 2 min. Primer concentration (300 pmol) was determined using QubitTM (ThermoFisher).
Colony PCR
Next, we did colony PCR of a single colony of E. coli. This was sterilely diluted in 300 µL autoclaved deionized water. A 1 µL aliquot was transferred to a 50 µL PCR reaction.
50 µL colony PCR reaction mix:
1 µL of forward primer (Kilobaser / eurofins)
1 µL of reverse primer (Kilobaser / eurofins)
1 µL of diluted single E. coli colony
25 µL of Dream Taq PCR master mix (Thermo Fisher)
Nuclease free water to 50 µL
Thermal cycling profile:
95°C for 1.5 min
38 times 95°C for 30 s, 53°C for 30 s, 72°C for 1 min
72°C for 10 min
The PCR fragments were then put onto a 1% agarose gel and separated according to size.
Results
We expected the DNA fragment length as followed:
UL30 – 179 bp
gC – 197 bp
UL6 – 142 bp
Figure 1 shows the resulting agarose gel on the transilluminator. Using the 100 bp standard ladder, we can see that UL30 is just below the 200 bp mark as expected. The DNA fragment matching the UL6 primers also shows the expected size. The gC fragments turned out to be much larger than expected, here we can assume a bad primer design or not matching annealing temperature. Nevertheless, there is a DNA fragment that at least indicates one binding primer. The PCR reactions show no difference in fragment length and intensity for all comparisons.
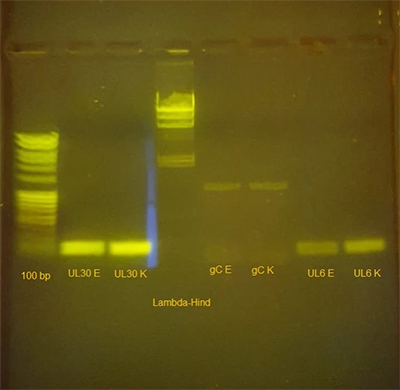
Figure 1: Agarose gel of a colony PCR for detection of artificial bacterial chromosomes in E. coli. Bands described from left to right: 100 bp ladder, UL30 with Eurofins(E) primers, UL30 with Kilobaser(K) primers, Lambda-Hind ladder, gC with Eurofins primers, gC with Kilobaser primers, UL6 with Eurofins primers, UL6 with Kilobaser primers.
There is no difference in fragment length and intensity between the PCR products performed with Kilobaser synthesized primers and the primers ordered from the online synthesis provider (Eurofins).
Conclusion
We have demonstrated that the selected bacterial colony contains the BAC. Therefore, the DNA from this colony can be extracted and sequenced.
The primer for the gC DNA fragment needs to be redesigned because at least one primer did not bind to the target sequence.
In addition, we have demonstrated that DNA primers synthesized with the Kilobaser personal DNA & RNA Synthesizer give the same results as primers ordered online from the synthesis provider.
Want to read more exciting articles in the future?
Are you interested in our new products and don’t want to miss a thing? Subscribe to our newsletter and stay up to date on what’s going on!

Share this article: