Fluorescent Probe – The Shining Star of PCR
Fluorescent probes have become more and more established in molecular biology research and development. Many scientists are working with these shining wonders of nature in fields such as medicine, pharmaceuticals and green biotechnology. Brush up on your knowledge of fluorescently labeled probes. The better informed you are, the better you can use fluorescent probes in your research.

DNA PROBE DEFINITION
The term ‚probes‘ is commonly used to describe fluorescent oligonucleotides. The most used DNA probes are dual-labeled and thus consist of three distinct parts:
a single strand DNA between 5 and 35 bp responsible for specific DNA recognition
a reporter dye emitting fluorescence at a certain wavelength
a quencher absorbing the light emitted by the reporter dye at a certain state.
IT’S ALL PHYSICS
DNA probes are labeled with an appropriate pairing of reporter dye and quencher, to utilize Förster resonance energy transfer (FRET for short).
FRET is a physical process of energy transfer named after Theodor Förster. In this process, the energy of a stimulated dye, also called a donor, is transferred to a second dye, also called an acceptor. However, energy transfer can only take place if acceptor and donor are in close proximity to each other. The energy is exchanged radiation-free and thus not via emission of light particles (photons).
In other words, FRET is a well-known quenching mechanism depending on the distance of emitting dye and absorbing quencher.
When we talk about DNA probes, the donor is called reporter and the acceptor is called quencher.
THE LIGHT SIDE
Let’s take a closer look at the fluorescent side.
Fluorescence is defined as ‚the spontaneous emission of light in response to irradiation by light‘. In other words, after optical energization of an electron, it enters a state of higher energy. However, this state is highly unstable.
As soon as the energized electron returns to the lower energy state, excess energy is emitted in the form of photons.
Fluorophores come in all colors of the rainbow, depending on the energy they emit.
SIZE MATTERS
For short DNA probes, the separation of reporter dye and quencher occurs after a polymerase has hydrolyzed the probe and released the reporter dye.
Probes that require hydrolysis are referred to as TaqMan probes, based on TaqMan polymerase with 3′-5′ exonuclease activity.
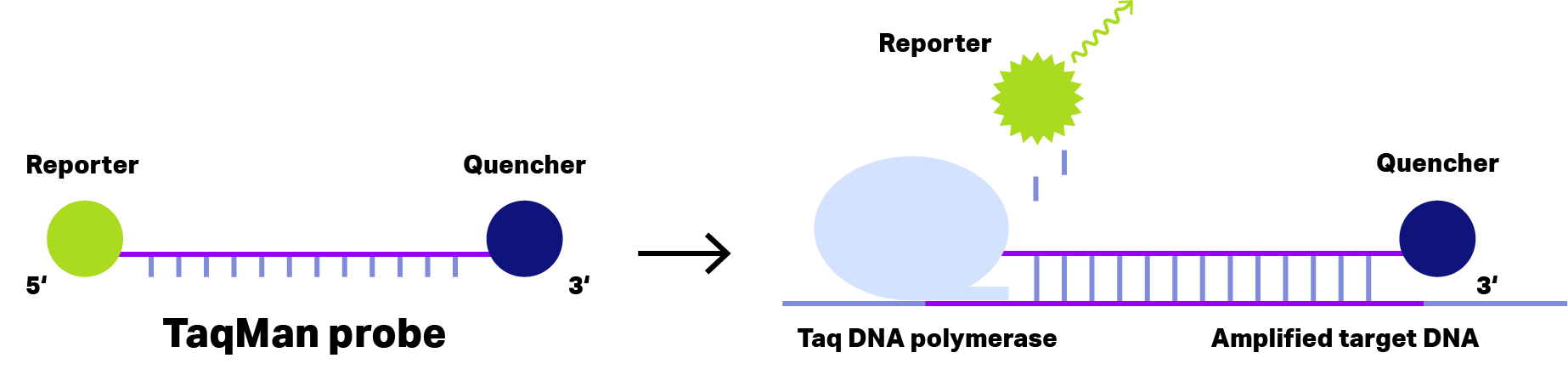
Figure 1: Principle of TaqMan Probe. In case of short dual-labeled oligos (left), the distance of reporter dye (green) and quencher (blue) is sufficient small to inhibit fluorescence emission, even after binding to the amplified target DNA. During the elongation phase of the PCR, Taq DNA polymerase with exonuclease activity cleaves the reporter dye from the oligo, allowing fluorescence emission detecting the target DNA..
Longer DNA probes form hairpin, loop or coil structures. With this structure, the quencher stays close enough to the reporter and quenches (absorbs) any emission.
However, as soon as probes bind on their target DNA, reporter and quencher get separated.
For these DNA probes, separation by binding to the target DNA is already sufficient to neutralize the quenching effect. Now, the emitted signal can be detected.
Probes emitting after binding are called hybridization probes.
The best-known hybridization probes are Molecular Beacon, Scorpion, Amplifluor® and LUXTM (see Figure 1)
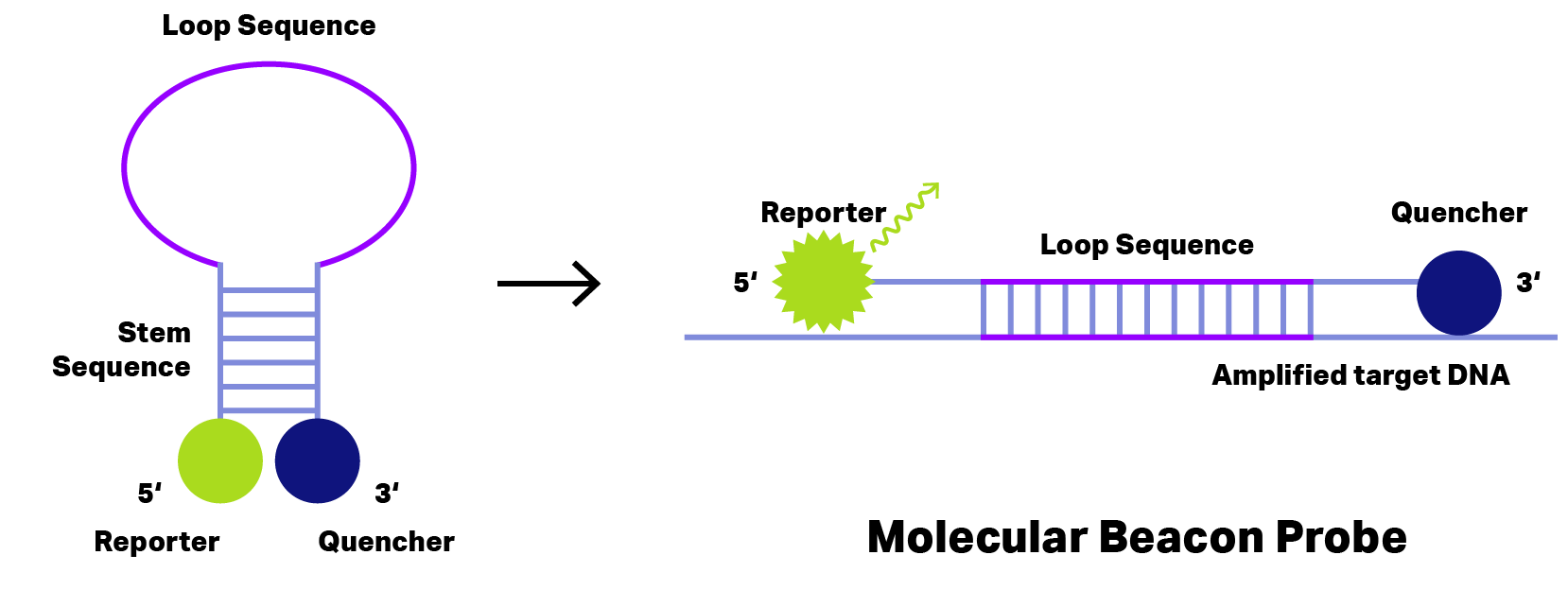
Figure 2: Principle of Molecular Beacon Probe. During hairpin formation (left) the quencher (blue) stays very close to reporter dye (green) and thus fluorescence emission is inhibited. Binding to the amplified target DNA (right) change the probe conformation separating reporter dye and quencher to allow fluorescence emission detecting the target DNA.
THE USE OF FLUORESCENT PROBES
One of the major uses of probes is to visualize targeted DNA sequences for quantitative real-time PCR (qPCR), which became an extremely versatile tool due to the fluorescent probes.
Quantitative PCR can also be performed with fluorescence dyes binding to dsDNA, whose amount increases during the PCR. However, these dyes bind to any dsDNA without distinguishing target DNA from other DNA strands. In contrary, probes show higher specificity due to specific oligo part annealing only on the target sequence.
Hybridization probes allow further applications, beside quantification of pathogens also mutation detection, melting point analysis and in situ hybridization.
Furthermore, probes can be labeled with different reporter dyes and thus allow multiplexing PCR to detect multiple targets in a single reaction.
THE STORY OF DUAL-LABELED FLUORESCENT PROBE SYNTHESIS
The first dual-labeled probes available for PCR were synthesized in two steps. At first a target specific oligo was synthesized with a common solid phase synthesis with phosphoramidites. Thereby, the last nucleotide was further modified with the reporter dye on its 5’-end.
After releasing the DNA oligo from the solid phase, the quencher was added by coupling an active ester to the 3′-end amino group. As the post-synthetic coupling of the quencher was not fully efficient, the obtained probes needed to be purified. Not only from incomplete oligos but also from probes, which were not quenched. Both, the post-synthetic modification, and the purification limited the evolving of probes for PCR initially.
In the mid-1990s, controlled pore glass (CPG) became available as synthesis support columns. CPG columns allowing solid state synthesis of oligos that start directly with a nucleotide 3′-end modification. For fluorescent labeled probes the columns are modified with a 3′-end quencher.
As of this date, the outcome of the common solid-phase synthesis is a dual-labeled oligo. Accordingly, we can say the introduction of CPG columns with 3′-end quencher simplified the synthesis of probes significantly.
COME TO THE DARK SIDE
Further improvement for fluorescent probes came in 2000 with the introduction of the first non-fluorescent quencher, Black Hole Quencher 1 (BHQ-1) by Biosearch Technologies.
Up to that point, a second fluorescent dye was used as a quencher. TAMRA was frequently used for this purpose. The problem with TAMRA is that absorption (557 nm) and emission (583 nm) are very similar in wavelength. This causes the signal emitted by the reporter to energize the quencher, which then emits an interfering background signal, resulting in low detection of the reporter signal.
The idea was to find a quencher dye which does not emit fluorescent signals by itself. A solution was found in BHQ with its polyaromatic-azo backbone offers the solution. Through this discovery, a wide variety of quenchers with a broad absorption range became available. In combination with FRET, the background signal is kept to a minimum for many reporter dyes.
BHQ-1 quenches in the range 480 – 580 nm and can be used in conjunction with the commonly used fluorophores, e.g. FAM, TET, JOE and HEX.
Beside the lack of intrinsic background fluorescence, the BHQ-1 quencher does not require changes to standard oligonucleotide synthesis procedures. Therefore, it was possible to complete the synthesis and deprotection of a dual-labeled FRET probe on a standard automated DNA synthesizer.
WELL PREPARED IS HALF THE BATTLE
Now you have an extensive background knowledge of DNA probes and can become a shining star in the lab yourself!
Kilobaser DNA & RNA desktop synthesizer helps you design and synthesize custom DNA/RNA probes as fast and easy as possible.
Alex shows you exactly how it works in our tutorial‚ how to synthesize DNA with Kilobaser.
You have already watched it? Then book your exclusive demo and get all your questions answered! We are looking forward to meeting you!
Want to read more exciting articles in the future?
Are you interested in our new products and don’t want to miss a thing? Subscribe to our newsletter and stay up to date on what’s going on!

Share this article: