One Step RT-qPCR – Application Note
With this application we demonstrate how to perform a one-step RT-qPCR with primers and probes synthesized with Kilobaser personal DNA & RNA synthesizer. Here, we perform a detection of SARS-CoV-2 using TaqMan probe based RT-qPCR. The results prove both primers and probes synthesized with Kilobaser are comparable to those of commercial kits in terms of sensitivity and linearity.

one-step RT-qPCR to detect Sars-CoV-2
Introduction
At present early detection of infection by SARS-CoV-2 relies on the efficient detection of the viral genome using reverse transcriptase quantitative PCR (RT-qPCR). [1-3] RT-qPCR is performed by specifically targeting the viral genome using complementary synthetic oligonucleotides (oligos), particularly at least: 2 primers and 1x TaqMan probe. As these oligos are sequence specific, viral genetic variants with changes to the primer/probe binding region may reduce the performance of PCR assays and have the potential to cause assay failure. [4,5] Therefore primers and probe need to be adapted continuously on the virus variants.
Kilobaser DNA & RNA synthesizer empowers everyone, everywhere to develop their own primer and probes for their RT-qPCR independently from oligo manufacturer. Therefore, we developed a DNA synthesizer allowing the subsequent synthesis of primer and probe. This synthesizer can be easily implemented in any molecular diagnostic laboratory allowing the development of new primer and probe sets for a highly mutagen pathogens such as SARS‐CoV‐2 without the risk of contaminations that can occur at manufactures.[6]
We aim here to show that primers and probes synthesized with the Kilobaser Personal DNA Synthesizer meet the requirements for testing new primers and probes for RT-qPCR development. We evaluated sensitivity with positive control transcripts and compared them to commercial kits.
TaqMan probe based RT-qPCR
In TaqMan probe-based PCR, a fluorescent probe is used to allow detection of specific target DNA that is amplified during PCR. An oligonucleotide is labeled with a fluorescent reporter dye (here 6-FAM) at the 5′ end and a quencher (here BHQ-1) at the 3′ end. While the probe is intact, the proximity of the quencher reduces the fluorescence emitted by the reporter dye through a process called fluorescence resonance energy transfer (FRET).
When the target sequence is present, the probe binds between the primer sites and is cleaved during primer elongation by the 5′ nuclease activity of the polymerase. This cleavage of the probe separates the reporter dye from the quencher, increasing the signal from the reporter dye. In addition, the probe is removed from the target strand, allowing primer elongation to continue to the end of the template strand. Thus, the entire PCR process is not affected by the installation of the probe.
With each cycle, additional reporter dye molecules are cleaved from their respective probes, resulting in an increase in fluorescence intensity. The signal increase is therefore proportional to the amount of amplified DNA. The higher the initial copy number of the nucleic acid target, the more likely a significant increase in fluorescence will be detected.
KILOBASER DEVICE DESCRIPTION
Kilobaser personal DNA & RNA synthesizer is a benchtop sized micro column synthesizer. Its technology is based on a reagent cartridge system coupled with microfluidic chip. The device allows 5′-modifications via cartridge and 3′-modifications via chip. A 5′-modification is attached to the DNA strand at the end of a synthesis, whereas a 3′-modification is attached to the micro column, and the DNA strand synthesis is thus attached to the modification.
Due to the minimal consumption of reagents during the synthesis of DNA products, the turnaround time is kept extremely low (2.5 min per baser + 30-50 min post processing).
To synthesize the DNA primer and TaqMan probe set used in this protocol, Kilobaser 6-FAM DNA cartridge, Kilobaser standard chips and Kilobaser BHQ-1 chips are required.

Material and Methods
Preparation of primer and probes
Following the CDC-developed assay we synthesized primer and probes that targets the Nucleocapsid gene of SARS-CoV-2 – N1 and N2 using Kilobaser personal DNA synthesizer. To synthesize the TaqMan probes and primers for this experiment, one 6-FAM DNA Cartridge, two BHQ-1 chips and 4 standard chips were used.
The primer and probe sequences for the N1 and N2 genes are below:
2019nCoV_N1-FP, GACCCCAAAATCAGCGAAAT
2019nCoV_N1-RP, TCTGGTTACTGCCAGTTGAATCTG
2019nCoV_N1-FAM, FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ-1
2019nCoV_N2-FP, TTACAAACATTGGCCGCAAA
2019nCoV_N2-RP, GCGCGACATTCCGAAGAA
2019nCoV_N2-FAM, FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ-1
After synthesis primer and probes were resuspended in 12 µL TRIS-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) by vortexing for 2 min. Primer concentration was determined using QubitTM (ThermoFisher). Probes were purified using the magnetic beads-based purification kit by Kilobaser. The concentration of probe in the final eluate was determined via NanoPhotometerTM (Implen) by measuring the absorption of BHQ-1 at 534 nm (e: 34000 cm-1M-1).
One-step RT-qPCR PCR
25 μL PCR reaction mix:
6.25 μL of 2019_nCoC_N positive control (IDT, 10006605) from a 10-fold dilution series
12.5 μL of 2x Luna Universal Probe One-Step reaction mix provided with Hot Start Taq DNA Polymerase (New England BioLabs)
1.25 μL of 20x Luna WarmStart® RT Enzyme Mix from the kit (New England BioLab)
5 µL of a master mix containing primer (500nM), probe (125nM) and nuclease free water.
The master mix was prepared with the freshly synthesized primer and probes or with the 2019n CoV kit by IDT (Catalog-Nr. 10006605).
Thermal cycling profile (using Open qPCR real-time thermocycler (Chai Bio)) :
55 °C for 15 min for reverse transcription
95 °C for 15 min for Hot Start TaqMan probe activation
45 cycles of 95 °C for 30 s, 60 °C for 60 s
Results
Amplification curve and Standard curve
The analytical sensitivity of the primer/probe sets for each target gene was assessed using a 10-fold dilution series of SARS-CoV-2 positive control starting from 10 000 copies down to 0.1 copies. From the resulting amplification curves the Ct-values were determined for each concentration except for the amplification curve at 0.1 copies target gene as the curve were not reproducible indicating the detection limit of the used conditions. With the calculated Ct-values linear regression was performed to obtain the slope and R2. The determined amplification curves and standard curve obtained with Kilobaser primer/probe sets are shown together with the resulting standard curves in Figure 1 and 2.
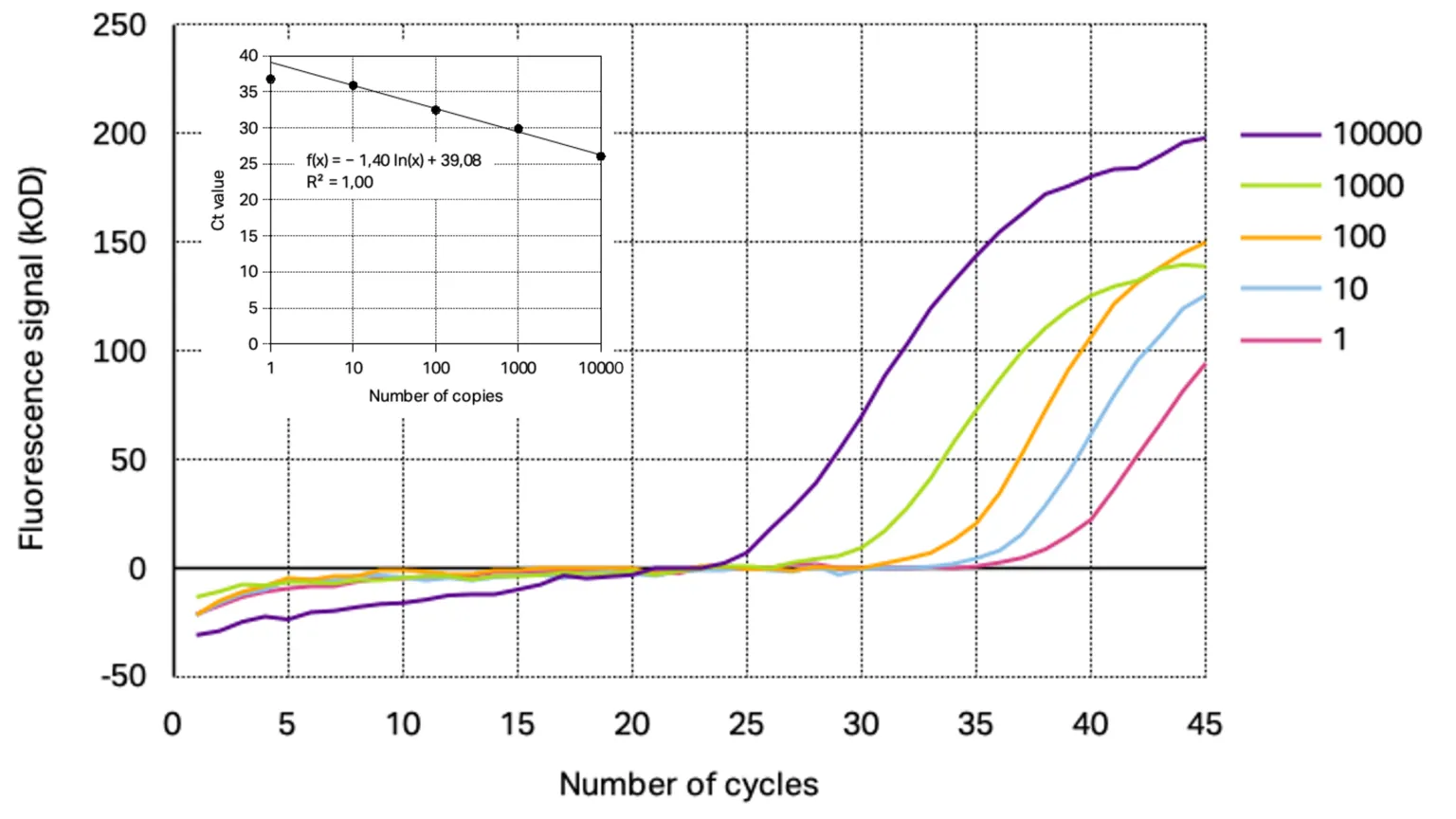
Figure 1: Representative amplification curves of RT-qPCR at different concentrations of 2019_nCoC_N positive control using Kilobaser primer and probes for N1 gene detection. The insert shows resulting standard curve with its determined slope and linear regression
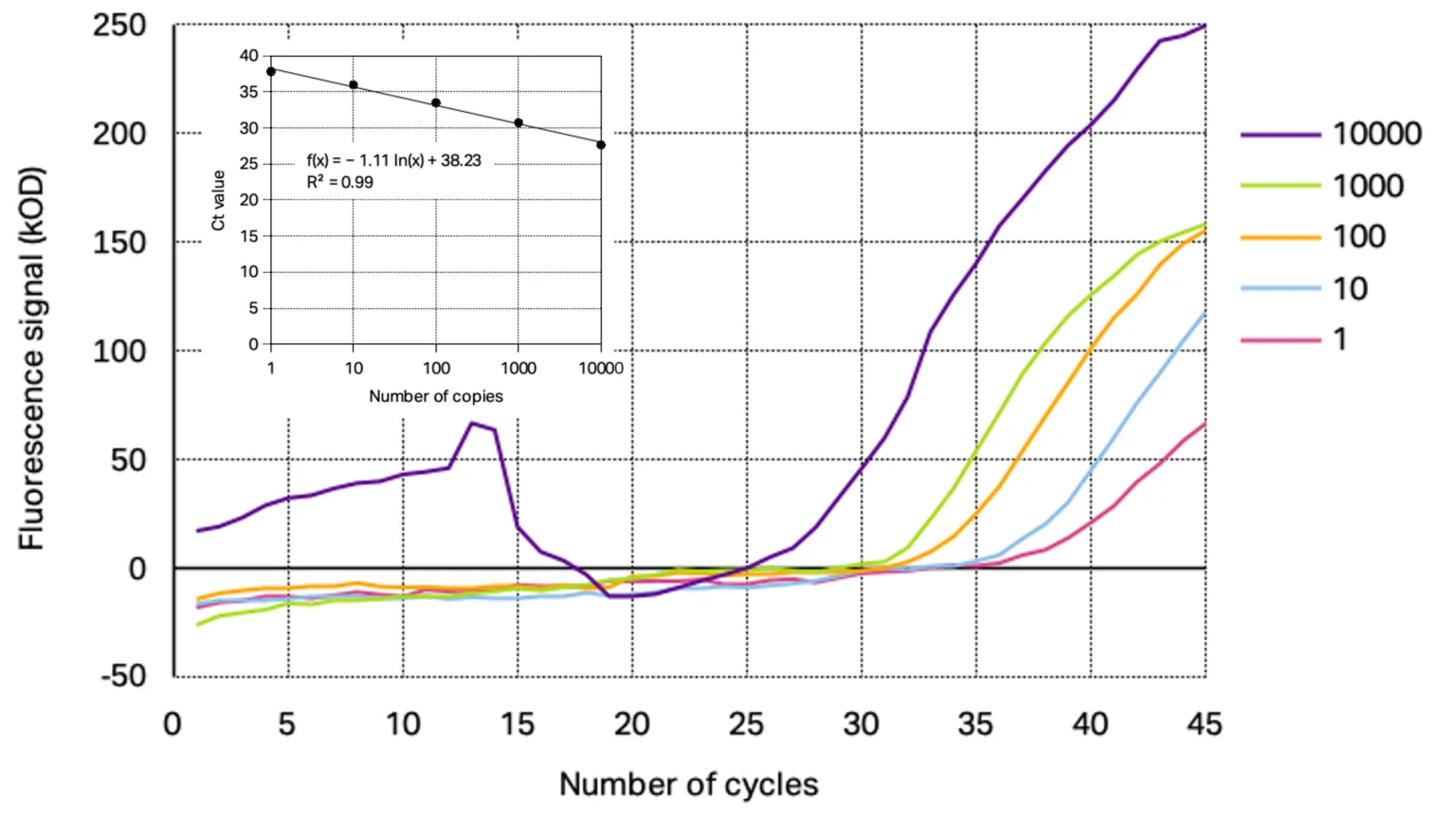
Figure 2: Representative amplification curves of RT-qPCR at different concentrations of 2019_nCoC_N positive control using Kilobaser primer and probes for N2 gene detection. The insert shows resulting standard curve with its determined slope and linear regression
Comparison with commercial primer/probe sets
Applying the commercial kit with ready to use primer/probe set at the same concentration as Kilobaser synthesized primer/probe set, similar amplification curves and standard curves were determined. The direct comparison of the curves, as seen in Figure 3, shows that even the progress as well as the signal intensity are nearly identically resulting in standard curves with similar slope and limit of detection.
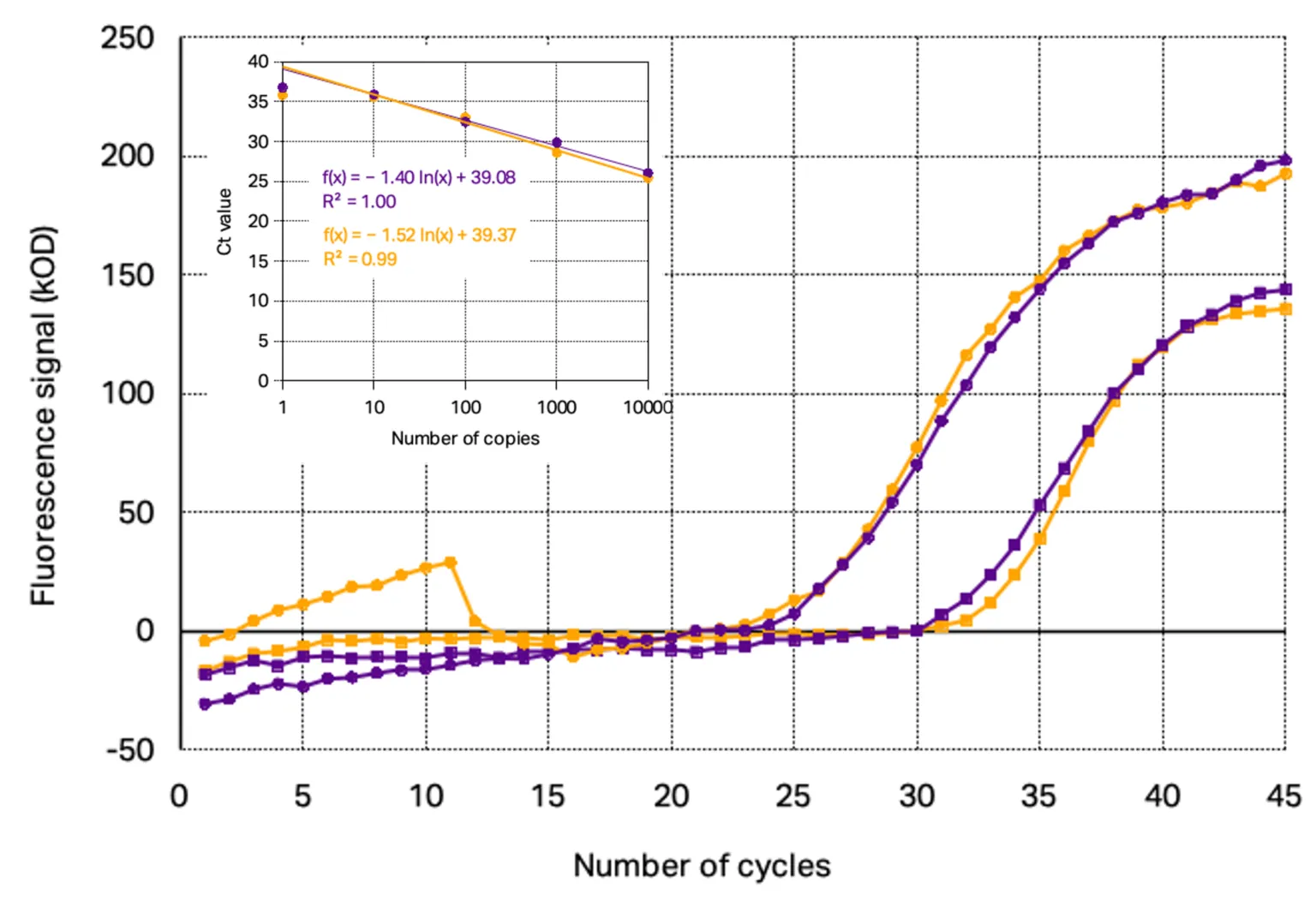
Figure 3: Comparisons of RT-qPCR of Kilobaser synthesized primer/TaqMan probe sets (purple) and ordered primer/TaqMan probe sets (orange): PCR amplification curves with 10 000 copies target gene (circles) and 100 copies target gene (squares) with their standard curves
The main difference that could be found is that the background signal of the TaqMan probes synthesized with Kilobaser personal DNA & RNA synthesizer showed with 520±39 kOD a significant higher signal at the beginning of the PCR as the commercial HPLC-purified probes with 360±17 kOD fluorescence signal. This observation indicates that Kilobaser synthesized probes feature a slightly lower quenching efficiency as the probes from oligo suppliers. However, since the background signal is still low, it does not affect the fluorescence measurement during PCR.
Conclusion
Based on our conditions, the primer/probe set synthesized with Kilobaser performed as well as the ordered commercial primer/probe set in terms of sensitivity and linearity. A similar standard curve was obtained with both primer/probe sets. This proves that the quality of the primers and probes synthesized by Kilobaser can also be used for sensitive qPCR measurements and is thus also suitable for the development of new primers and probes for PCR-based detection.
1. Daniella Parilli-Troconis, Peter Baptista, Marcel Marcano-Lozada, Stefania Goncalves, David Shahal, Juan Armando Chiossone-Kerdel. COVID-19 Infection and Its Influence in Otorhinolaryngology-Head and Neck Surgery. Int Arch Otorhinolaryngol 2020; 24(04): e527-e534.
*DOI: 10.1055/s-0040-1715586. *
*2. Nandini Sethuraman, Sundararaj Stanleyraj Jeremiah, Akihide Ryo. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA. 2020;323(22):2249-2251. DOI:10.1001/jama.2020.8259. *
*3. Nadin Younes, Duaa W. Al-Sadeq, Hadeel AL-Jighefee, Salma Younes, Ola Al-Jamal, Hanin I. Daas, Hadi. M. Yassine, Gheyath K. Nasrallah. Challenges in Laboratory Diagnosis of the Novel Coronavirus SARS-CoV-2. Viruses 2020, 12(6), 582; *
*4. Nathaniel Storey, Julianne R Brown, Rui P A Pereira, Denise M O’Sullivan, Jim F Huggett, Rachel Williams, Judith Breuer, Kathryn A Harris. Single base mutations in the nucleocapsid gene of SARS-CoV-2 affects amplification efficiency of sequence variants and may lead to assay failure. Journal of Clinical Virology Plus 1 (2021) 100037. *
*5. Pillonel Trestan, Scherz Valentin, Jaton Katia, Greub Gilbert, Bertelli Claire. Letter to the editor: SARS-CoV-2 detection by real-time RT-PCR. Euro Surveill. 2020;25(21):pii=2000880. *
*6. Claire Y. T. Wang, Cameron Buckley, Cheryl Bletchly, Patrick Harris, David Whiley. Contamination of SARS-CoV-2 RT- PCR probes at the oligonucleotide manufacturer. Pathology (2020), 52(7), December. *
You want to learn more about Kilobaser?
You are only 3 steps away from your ready-to-use DNA. Take a closer look and let us explain all the details in your exclusive demo!

Share this article: