CRISPR/Cas9 – Application Note
This application demonstrates how to perform a CRISPR/Cas9 cleavage using a Kilobaser synthesizer. For cleavage, an sgRNA was transcribed from a DNA template prepared using the Kilobaser Synthesizer and Purification Kit.

Efficient sgRNA synthesis for CRISPR/Cas9
by Sabine Sykora. Ph.D., MABEAL GmbH
Abstract
The specificity and simplicity of the RNA-guided cutting system CRISPR/Cas9 make it a widely used gene editing tool. To use this cutting system for in vitro application, the single guide RNA (sgRNA) is typically prepared by transcribing a DNA template into the fully functional sgRNA. Today, one-step in vitro transcription kits are available that allow the preparation of sgRNA in 30 minutes. The DNA template for this kit can be synthesized with any commercially available DNA synthesizer.
As the developer of our own DNA synthesizer, we tested the application ourselves. We synthesized the DNA template, transferred it to the sgRNA and used it to cut a plasmid with the Cas9 nuclease. We found that the enzymes used in the transcription kit transcriptase and DNase are inhibited by residues from the synthesis.
After removing these residues with our OliPure HIC purification kit, the same yield and functionality of sgRNA was obtained with Kilobaser DNA template as with DNA template from an oligo supplier. This result proves that by using Kilobaser one/one-XT and OliPure HIC Kit in combination with a transcription kit, any lab can use CRISPR/Cas9 for gene editing.
|
Device: | |
|
Cartridge: | |
|
Chip: | |
|
Supply: | |
|
Supplementary: |
EnGen® sgRNA Synthesis Kit by NEB Monarch® RNA Cleanup Kit by NEB |
INTRODUCTION
CRISPR/CAS9 CUTTING SYSTEM
Since its discovery by Emmanuelle Charpentier and Jennifer A. Doudna [1], the CRISPR/Cas9 system has been adopted as a widely used toolkit for gene editing in vivo and in vitro [2-6]. This is achieved by combining a single guide RNA (sgRNA) with the endonuclease Cas9. The latter is guided to the desired cleavage site on the target DNA by the complementary sequence of the sgRNA, making the nuclease activity programmable with the sequence of the sgRNA[1-7].
The sgRNA is a version of the naturally occurring two-part guide RNA complex engineered into a single sequence. It consists of the native crRNA, which guides Cas9 to the appropriate target site, and a trans-activating crRNA (tracrRNA), which provides a scaffold for Cas9 binding. [7-9]
THE SGRNA OF THE CAS9 NUCLEASE IS COMPOSED OF TWO FUNCTIONAL UNITS:
crRNA: a targeting region containing a 20nt sequence complementary to the DNA target sequence. [7,8]
tracrRNA: a scaffold region consisting of three stem loops that complex with the Cas9 nuclease. [8,9]
To cut a target double-stranded DNA (dsDNA), the sgRNA first binds to the Cas9 nuclease, forming the Cas9-sgRNA complex. Upon complexing, Cas9 is directed into an active conformation that interrogates the next dsDNA strand to find the 3bp NGG protospacer adjacent motif (PAM). [8] Upon binding the PAM, the dsDNA is unwound, allowing the sgRNA to anneal to one of the DNA strands. If the DNA sequence near the PAM is sufficiently complementary to the crRNA sequence of the sgRNA, Cas9 cleaves the DNA target, producing a blunt-ended double-strand break. [7,8,10-13] This cleavage mechanism of the CRISPR/Cas9 system is illustrated in Figure 1.
For the cleavage two distinct domains are actually responsible [8,12]:
RuvC domain, which cleaves non-complementary DNA strands
HNH domain, which cleaves complementary DNA strands

Figure 1: DNA cleavage by CRISPR/Cas9 scissors. sgRNA, consisting of crRNA and tracrRNA, positions and activates the Cas9 nuclease on the genomic DNA. The correct position of Cas9 is determined by the binding of crRNA to the DNA strand complementary to the target DNA and PAM. Near the PAM, the two nuclease domains, RuvC and HNH, cut each of the DNA strands, creating a double-stranded break.
HOW DO WE GET THE SGRNA INTO OUR LAB?
The applicability of CRISPR/Cas9 depends on the availability of the sgRNA. However, working with such long RNA oligonucleotides is quite challenging due to their stability and synthesis. On the one hand, RNA synthesis is much more complex than DNA synthesis, making it difficult and expensive to synthesize. On the other hand, there is the sensitivity to environmental RNases, which reduces the lifetime of these oligonucleotides. Therefore, for a long time, sgRNAs have only been synthesized in vivo, directly in a cell. Meanwhile, sgRNAs can also be easily synthesized in vitro using in vitro transcription (IVT) kits in less than one hour, such as the EnGen sgRNA Synthesis Kit from New England Biolabs (NEB). [14]
In these kits, a synthesized DNA oligonucleotide containing the target-specific crRNA sequence is transcribed into a fully functional sgRNA. Three different steps can be distinguished as shown in Figure 2:
1. Oligo hybridization: A custom DNA oligonucleotide containing the target-specific crRNA sequence hybridizes with another DNA oligonucleotide containing the tracRNA sequence.
2. dsDNA extension: The hybridization is the starting point for a DNA polymerase to generate a double-stranded DNA (dsDNA) template for transcription.
3. Transcription from DNA to RNA: Immediately after the synthesis of the dsDNA template, fully functional sgRNA is amplified from it by an RNA polymerase.

Figure 2: Transcription of target-specific DNA oligonucleotide to functional sgRNA.
These kits are only half the solution. The target-specific DNA oligonucleotide or DNA temlate has to be synthesized first. The result of the sgRNA synthesis and thus the efficiency of the cleavage depends on the quality of the DNA template. The in-house synthesis of this oligonucleotide can be easily performed using our Kilobaser one.
To demonstrate the compatibility of our synthesis product with the EnGen sgRNA Synthesis Kit, we synthesized a DNA template to prepare an sgRNA that guides a Cas9 nuclease to the cleavage site on the target DNA, a plasmid.
PREPARATION OF SGRNA
DESIGN OF THE DNA TEMPLATE
The sgRNA was designed so that the plasmid pTZ19R is cleaved by the Cas9 of S. pyogenes (SpCas9) near the cutting site of the restriction enzyme Sac I. The use of either of these endonucleases changes the form of the plasmid from open circular to linear, which can be detected by gel electrophoresis because the linear form moves faster than the circular form.
The PAM sequence in the target is crucial for the cutting site of CRISPR/Cas. Therefore, the sequence of the 20nt crRNA was determined from the target sequence by searching for a PAM sequence close to the desired cleavage site.
This sequence was inserted between the T7 promoter sequence (upstream) and an overlap with the SpCas9 specific scaffold (downstream) to form the 55nt target-specific DNA oligonucleotide for the sgRNA synthesis kit as shown in Figure 3.
As recommended by the NEB protocol for target-specific oligo design an initial 5’-dG was added to the target-specific sequence to enhance the binding of the T7 RNA polymerase transcription.

Figure 3: Design of target-specific DNA consisting of the T7 promoter sequence (left), the target-specific crRNA sequence with an initial 5’-dG (middle) and the overlap with the SpCas9 specific scaffold (right).
PREPARATION OF THE DNA TEMPLATE
The DNA template for the sgRNA synthesis kit was synthesized using Kilobaser one with a standard chip and standard DNA cartridge. After synthesis, the DNA template was resuspended in 12μL TRIS-EDTA buffer (10mM Tris-HCl, 1mM EDTA, pH 8.0) by vortexing for 2 min.
The resulting suspension contains the DNA template as well as salts from the protecting groups, some truncated oligonucleotides and unreacted phosphoramidites. These residues from hydrophobic synthesis reagents can affect enzymes by binding to their substrate binding site and inhibiting the enzymatic reaction.
To detect this inhibition, the synthesized DNA template was used either directly or after removal of synthesis residues by purification with the OliPure HIC Kit (Kilobaser). In addition, the sequence of the DNA template was ordered from an oligo supplier, which arrived desalted, which is of the same quality as our purified DNA template.
The concentration of purified and unpurified DNA oligonucleotides was determined using an Invitrogen™ Qubit™ 3 Fluorometer.
SGRNA SYNTHESIS
The DNA template was transcribed in vitro using EnGen® sgRNA synthesis kit (NEB) according to manufacturer’s instructions.
The sgRNA synthesis was performed by mixing the DNA template with the kit reagents as follows: 10μL EnGen® sgRNA reaction mix (dNTPs, S. pyogenes Cas9 scaffold oligo); 0.6μL DNA template (9.5μM); 1μL DTT (0.1M); 2μL EnGen® sgRNA enzyme mix, made up to 20μL with nuclease-free water. The mixture was incubated at 37°C for at least 30 minutes.
Then 1μL of DNase I was added and incubated at 37°C for 15 minutes to digest the DNA template. The reaction mixture was then purified using the Monarch® RNA Cleanup Kit
(NEB). The resulting sgRNAs were stored at -20°C or used immediately for in vitro cleavage reactions.
After purification and before use, the sgRNA concentration was determined by measuring the absorbance at 260nm (A260) on the Implen NanoPhotometer® NP80 microvolume photometer using the following formula: A260 x 40 = „x“ μg/ml ssRNA. The molar concentration was determined using the molecular weight of 32794.6g/mol of sgRNA.
The final yields of sgRNA obtained with the different DNA templates were determined and are compared in Figure 4. The numbers show that the expected yields of 4-25μg were obtained with each DNA template. However, the unpurified DNA template from Kilobaser showed the lowest yield, while the same yield was obtained with both purified DNA templates.
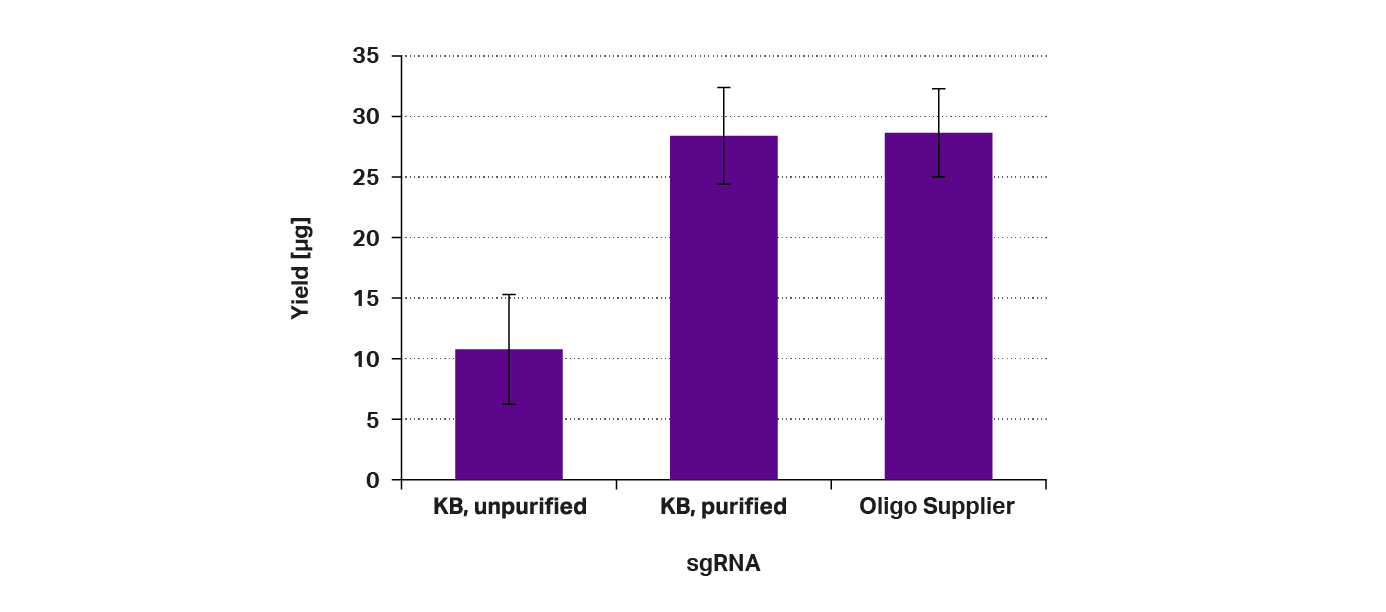
Figure 4: Concentration of sgRNA after purification – The sgRNA were transcribed from different DNA templates: (1) synthesized by Kilobaser (KB), unpurified; (2) synthesized by Kilobaser (KB), purified; (3) synthesized by oligo supplier, purified;
Based on these values, we can conclude that the residues from the synthesis do indeed inhibit the enzymes during sgRNA synthesis. After all, there are two enzymes, the DNA polymerase and the RNA polymerase, that are needed for the proper synthesis and amplification of the sgRNA.
DETECTION OF CAS9 ACTIVITY
The function of the synthesized sgRNAs was evaluated by cleaving the targeted plasmid with the Cas9-sgRNA complex and observing the linearization of the plasmid by gel electrophoresis.
PLASMID CUTTING
The reactions were set up as follows. First, the Cas9-sgRNA mixture was prepared: Cas9 enzyme (20μM, NEB) and sgRNAs were mixed in equimolar amounts in NEB 3.1 buffer (10x, NEB) and incubated at room temperature for 10 minutes. The plasmid (0.5 μg/μl, ThermoFisher) was then added and digested at 37°C for 30 minutes. To stop the cleavage and remove the Cas9 nuclease from the plasmid, the reaction mixture was incubated with 1μL Proteinase K (20mg/ml, CarlRoth) at room temperature for 10 minutes.
The stopped cleavage reaction was mixed with Gel Loading Dye, Purple (6x, Carl Roth). Together with 1kb Plus DNA Ladder (50μg, Carl Roth), the cleavage products were resolved on a 2% TBE agarose gel stained with SYBR™ Safe.
In the resulting gel, two different bands can be observed, representing the two different forms of the plasmid:
1. Open circular form: As long as the plasmid is not cleaved, it remains a closed circular DNA strand.
2. Linear form: It is the result of cleavage on both DNA strands by an endonuclease.
The migration of circular and linearized plasmids is affected by the compactness of each conformation as they move through the gel pores. The flexible linear form will migrate at a faster rate than the floppy open circular form and will therefore appear further down in the gel. This differential migration allows the separation of the cleaved and uncleaved plasmid.
Cleavage with the respective sgRNAs resulted in the following bands on the gel in Figure 5:
The uncleaved plasmid is found at the top (near the sample ridge) because it hardly migrated into the gel, while the cleaved products are represented by a band significantly deeper in the gel. The band is about 2.5kb, representing the length of 2862bp of the plasmid.
The cleavage approaches with sgRNA prepared with purified DNA template show mainly the second band at about 2.5 kb, indicating the ring opening of the plasmid and thus the cleavage process that has occurred. Uncleaved plasmid is barely detectable. This is the case for DNA template from Kilobaser and from the oligo supplier.
The cleavage approaches with the sgRNA prepared with unpurified DNA template also showed the band at about 2.5kb, indicating cleavage. However, significantly more uncleaved plasmid was detected than cleaved plasmid.
No cleavage product was detected in the control mixtures containing either sgRNA alone or Cas9 alone, indicating effective targeting to the desired cleavage site by all synthesized sgRNAs.
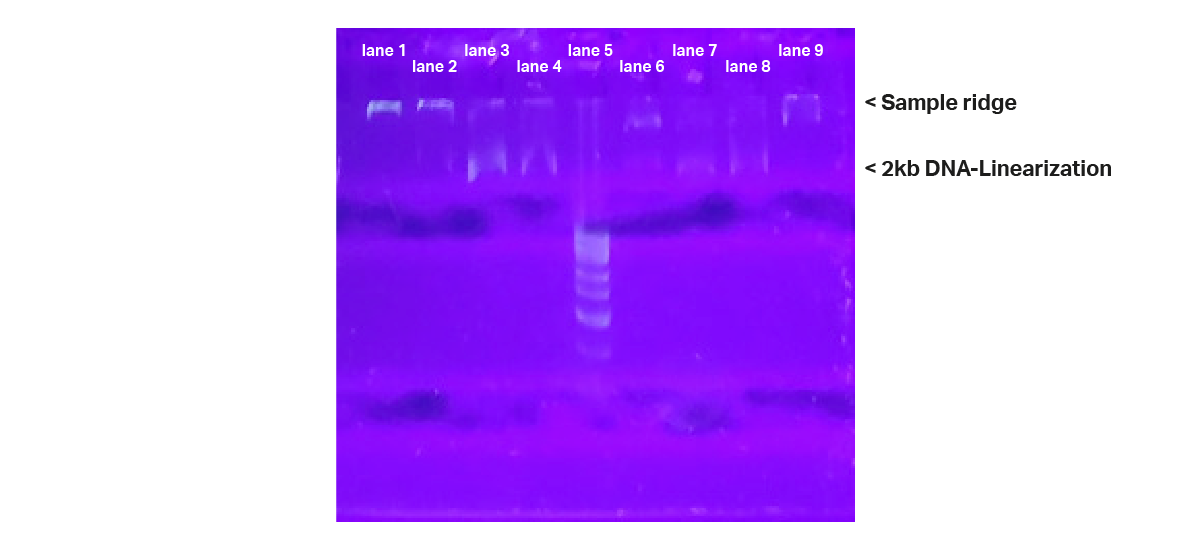
Figure 5: Gel-Electrophoresis. Gel shows duplicate lanes for each sample. lane 1: no Cas9 control; lane 2+6: sgRNA Kilobaser, unpurified; lane 3+7: sgRNA Kilobaser, purified; lane 4+8: sgRNA oligo supplier, purified; lane 5: DNA ladder; lane 9: no sgRNA control; Cleavage products at expected band sizes were detected for all samples containing the sgRNA + Cas9 + plasmid. No product at the expected band size was observed for all the control samples.
EFFECT OF PURIFICATION ON ACTIVITY
Gel electrophoresis results (Figure 5) showed significant differences in nuclease activity whether the DNA template was purified or not. This observation is interesting because neither the concentration nor the quality of the sgRNAs should be different. The lower yield of the sgRNA synthesis was compensated by using more sgRNA. Due to the purification after the sgRNA synthesis, there were no synthesis residues left to interfere with the nuclease activity. There are two possible explanations for this observation.
If the concentration of sgRNA was the same for all cutting reactions, the difference in activity could be due to errors in the transcription process. Under the influence of the synthesis residues, the RNA polymerase may have transcribed incorrectly, adding base mismatches. These mismatches would affect the recognition of the target site and thus the cutting efficiency.
It is also possible that the concentration of the sgRNA was not the same. The concentration was determined by measuring the absorbance at 260nm. This method does not distinguish between RNA and DNA. After sgRNA synthesis, the DNA template for transcription is degraded by the added DNase. This degradation occurs before purification, so it is possible that the DNase was inhibited by the synthesis residues. As a result of the inhibited DNA degradation, the DNA remains side by side with the RNA oligonucleotide even after purification, affecting the concentration determination. In the end, a false high concentration of sgRNA was measured, resulting in a false low concentration for the cleavage assay.
Inhibition of DNase is more likely to be the cause of the low nuclease activity than transcription errors. With this observation, we found another reason to purify the DNA template after synthesis.
Post-synthesis purification is typically not required for short oligonucleotides including primers for PCR. However, in the final step of synthesis, protective groups are released from the oligonucleotide with each base. The amount of protective groups increases with each additional base. Consequently, synthesis of the 55nt long DNA template results in a higher amount of protective groups. These groups at this high concentration will interfere with any enzyme during sgRNA synthesis and therefore must be removed to obtain functional sgRNA
CONCLUSION
Since the applicability of CRISPR/Cas9 depends on the availability of sgRNA, it is important to have this oligonucleotide in the right quality at the right time. Therefore, in-house synthesis is a good solution for research labs that frequently use CRISPR/Cas9.
Our Kilobaser one synthesizer quickly and easily produces target-specific DNA oligonucleotides for use as DNA templates in available in vitro transcription kits. It is essential for the use of DNA oligonucleotides in these kits that the oligonucleotides are purified from synthesis residues, as these residues interfere with the enzymes used in these kits. The purification is easily performed using the OliPure HIC Kit.
Our tests show that purified Kilobaser DNA oligos are of the right quality to provide functional sgRNA for use in the CRISPR/Cas9 cutting system. Therefore, the Kilobaser one DNA Synthesizer is a good candidate for in-house synthesis for research using CRISPR/Cas9.
1. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012 Aug 17;337(6096):816-21.
doi: 10.1126/science.1225829. Epub 2012 Jun 28. PMID: 22745249; PMCID: PMC6286148.
2. Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013 Jan 29;2:e00471. doi: 10.7554/eLife.00471. PMID: 23386978; PMCID: PMC3557905.
3. Karvelis T, Gasiunas G, Siksnys V. Programmable DNA cleavage in vitro by Cas9. Biochem Soc Trans. 2013 Dec;41(6):1401-6. doi: 10.1042/BST20130164. PMID: 24256227.
4. Trevino AE, Zhang F. Genome editing using Cas9 nickases. Methods Enzymol. 2014;546:161-74.
doi: 10.1016/B978-0-12-801185-0.00008-8. PMID: 25398340.
5. Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014 Nov 28;346(6213):1258096. doi: 10.1126/science.1258096. PMID: 25430774.
6. Wang SW, Gao C, Zheng YM, Yi L, Lu JC, Huang XY, Cai JB, Zhang PF, Cui YH, Ke AW. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol Cancer. 2022 Feb 21;21(1):57.
doi: 10.1186/s12943-022-01518-8. PMID: 35189910; PMCID: PMC8862238.
7. Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):E2579-86.
doi: 10.1073/pnas.1208507109. Epub 2012 Sep 4. PMID: 22949671; PMCID: PMC3465414.
8. Jiang F, Doudna JA. CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys. 2017 May 22;46:505-529. doi: 10.1146/annurev-biophys-062215-010822. Epub 2017 Mar 30. PMID: 28375731.
9. Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011 Mar 31;471(7340):602-7. doi: 10.1038/nature09886. PMID: 21455174; PMCID: PMC3070239.
10. Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology (Reading). 2009 Mar;155(Pt 3):733-740
doi: 10.1099/mic.0.023960-0. PMID: 19246744.
11. Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014 Mar 6;507(7490):62-7.
doi: 10.1038/nature13011. Epub 2014 Jan 29. PMID: 24476820; PMCID: PMC4106473
12. Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014 Feb 27;156(5):935-49.
doi: 10.1016/j.cell.2014.02.001. Epub 2014 Feb 13. PMID: 24529477; PMCID: PMC4139937.
13. Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014 Sep 25;513(7519):569-73.
doi: 10.1038/nature13579. Epub 2014 Jul 27. PMID: 25079318; PMCID: PMC4176945.
14. Instruction manual for EnGen® sgRNA Synthesis Kit, S. pyogenes, https://international.neb.com/applications/genome-editing/in-vitro-transcription-of-sgrna
Want to read more exciting articles in the future?
Are you interested in our new products and don’t want to miss a thing? Subscribe to our newsletter and stay up to date on what’s going on!

Share this article: