Real Time qPCR – Application Note
Using COVID-19 detection as an application example, we performed a real time qPCR. To demonstrate the performance of Kilobaser DNA oligos, primers synthesized by Kilobaser were compared to primers purchased from Eurofins. The Cq values show that the primers from Kilobaser and Eurofins lead to the same result.
real time qpcr to detect covid-19 infection
COVID-19 pandemic impact in science
The COVID-19 pandemic, caused by the SARS-CoV-2 virus, has resulted in more than 510 million confirmed cases worldwide with over 6.5 million deaths by April 28, 2022. Billions of people on this planet live and work in disrupted conditions due to various forms of social distancing and frequent lockdown regulations. Science and research continue to suffer from interrupted supply chains and long delivery times of essential resources.
In particular, DNA oligo availability is approaching its limit due to the high demand for COVID-19 primers. Scientists have to expect unusually long waiting times to get their required DNA products.
Kilobasers’ mission is to democratize DNA and provide an independent and fast method for scientists around the world to obtain DNA and RNA oligos on equal terms.
Principle of real time qPCR
Nucleic acid amplification and detection techniques are used in a broad range of applications in all areas of life science – basic research, medicine, diagnostics, biotechnology, forensics and more.
In traditional PCR an amplified DNA product is detected in an end-point method. In qPCR the accumulation of DNA is monitored in real time as the reaction progresses during amplification. This can be done using a sequence-specific DNA probe with a fluorophore and a quencher or a fluorescent dye. In this experiment a fluorescent dye is used.
Real-time PCR connects amplification and detection into one single step. It can be used for qualitative (the presence or absence of a sequence) and quantitative (copy number) analysis. Within this experiment, a fluorescent dye is used to label the PCR product. The PCR instrument can monitor the accumulation of fluorescent signal during the exponential phase of the reaction for precise, sensitive and quantitative analysis of the PCR product. The measured fluorescence is proportional to the total amount of amplified DNA product or also called amplicon.
CQ value
Initially, the fluorescent signal, is too low to be detected, even though product accumulates. Eventually, when enough product is amplified, the fluorescent signal is detectable. The cycle number at which fluorescence can be detected is the basic result of qPCR and is called Quantification Cycle (Cq for short).
For example, low Cq values represent higher initial amount of the target DNA. Sometimes Cq values are also called Ct (threshold cycle) values or Cp (crossing point) values in literature. However, there is no difference in the meaning of these values. “Cq or Quantification cycle” is the correct naming according the MIQE guidelines as described in Bustin et al.; Clinical Chemistry 55:4; 2009.
KILOBASER DNA synthesis
Kilobaser personal DNA & RNA synthesizer technique is based on an innovative cartridge and microfluid chip system.
The closed synthesis system provided by the microfluidic chip reduces the required reagent volume to a minimum.
This makes it possible to synthesize DNA oligos in a short time with a cycle time of 2.5 min per base. In addition, standard DNA primers do not need to be purified after synthesis with Kilobaser, as the salt residues are negligible due to the low reagent consumption. The primers are dissolved in water after synthesis and can be used immediately.
For synthesis of the DNA primers used in this application, one Kilobaser standard DNA cartridge and 2 Kilobaser standard chips were used.

Material and Methods
Preparation of DNA primer with Kilobaser
Following the CDC-developed assay we synthesized primer and probes that targets the Nucleocapsid gene of SARS-CoV-2 – N1 using Kilobaser personal DNA synthesizer. To synthesize the primers for this experiment, one Standard DNA Cartridge and 2 standard Chips were used.
The primer sequence for SARS-CoV-2 – N1 gene detection:
2019nCoV_N1-FP, GAC CCC AAA ATC AGC GAA AT
2019nCoV_N1-RP, TCTGGTTACTGCCAGTTGAATCTG
After synthesis the primers were resuspended in 12 µL TRIS-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) by vortexing for 2 min. A total yield of 300 pmol was quantified with a fluorometer. Probes were purified using the magnetic beads-based purification kit by Kilobaser. The coupling efficiency of Kilobaser oligos is between 99.0% and 99.5%. Due to the low reagent volumes for synthesis with Kilobaser, there are negligible amounts of salt residues in the synthesis product, so no desalting is necessary.
real time qPCR
qPCR reaction mix:
1µl Template DNA (2019-nCoV_N Positive Control (IDT) (200.000 copies/µl) (IDT)
12.5 μL 5x Hot Firepol EvaGreen qPCR Mix Plus no ROX (Solis BioDyne
10 pmol Forward Primer (Kilobaser or Eurofins)
10 pmol Reverse Primer (Kilobaser or Eurofins)
Nuclease free water
The master mix was prepared with the freshly synthesized primer and probes or with the 2019n CoV kit by IDT (Catalog-Nr. 10006605).
Thermal cycling profile (using Open qPCR real-time thermocycler (Chai Bio)):
15 min 95°C initial denaturation
40 times:
15 sec 95°C
20 sec 50°C
60 sec 72°C5 min 70°C
Cq values were measured by the qPCR instrument (ChaiBio)
Results
The purple amplification curve in figure 1 shows a reaction where Eurofins forward and Kilobaser reverse primer were used. The pink curve indicates the use of Eurofins forward and reverse primer. The dark blue curve represents the reaction of Kilobaser forward and Eurofins reverse primer and the light blue curve shows the use of forward and reverse primer made by Kilobaser. The second values of the double determination were not shown due to clarity/illustration reasons.
The Cq value represents the initial amount of template present at the start of the amplification reaction. For example, low Cq values represent higher initial amount of the target DNA.
The measured values vary from 24.39 to 23.47, which means they are all in the same range. The Cq value of the curve where only Kilobaser primers were used is 24.39; the Cq value of the reaction where Eurofins primers were used, is smaller by the value of 0.39.
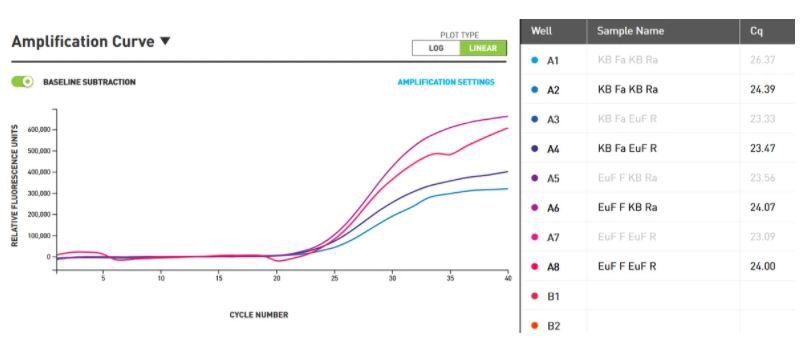
Figure 1: qPCR amplification curves performed by ChaiBio qPCR thermocycler. X-axis shows the cycle number and the y-axis shows the relative fluorescence units. Purple curve: forward primer (Eurofins) + reverse primer (Kilobaser). Pink curve: forward primer (Eurofins) + reverse primer (Eurofins). Dark blue curve: forward primer (Kilobaser) + reverse primer (Eurofins). Light blue: forward primer (Kilobaser) + reverse primer (Kilobaser). Baseline subtraction is active, meaning that the background signal is subtracted.
Conclusion
Comparing the Cq values resulting from the qPCR amplification, primers made by Kilobaser show the same performance as primers purchased from the online synthesis provider Eurofins. We have thus demonstrated that DNA primers produced with Kilobaser personal DNA & RNA synthesizers are of comparable quality to Eurofins DNA primers. This means Kilobaser is a supply chain independent and faster alternative to obtain DNA oligos for real time qPCR.
You want to learn more about Kilobaser?
You are only 3 steps away from your ready-to-use DNA. Take a closer look and let us explain all the details in your exclusive demo!

Share this article: